
Empanada version 1.1 Napari plugin out now.
The empanada-napari plugin is designed to make deep learning image segmentation more accessible for researchers working with electron microscopy (EM). Included in this plugin is MitoNet, a versatile model specifically tailored for segmenting mitochondria instances.
LEARN MOREOur paper on MitoNet, our generalist deep learning model that allows mitochondrial segmentation from any EM or volume EM dataset, is on the cover of Cell Systems. MitoNet was developed to help overcome the segmentation bottleneck in vEM workflows. See the publications tab for details.

Using MitoNet and Empanada, researchers can segment tens of thousands of mitochondria in 3D within a matter of hours instead of years. Here, 15,180 mitochondria from a kidney tissue vEM image volume were segmented in < 2 hours on a single GPU 16 GB RAM workstation.
“See publications tab for details”
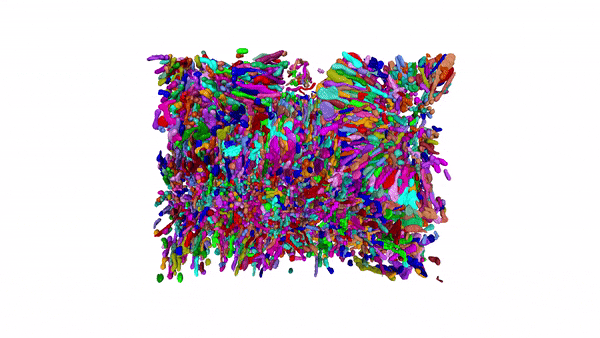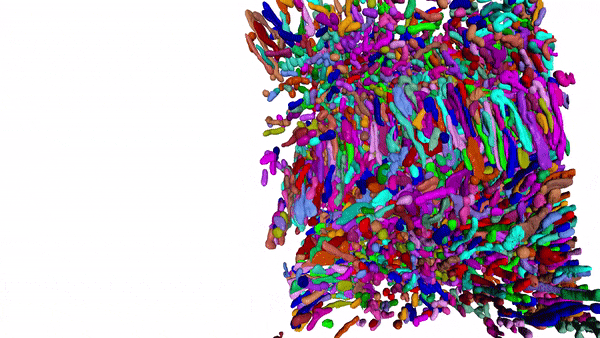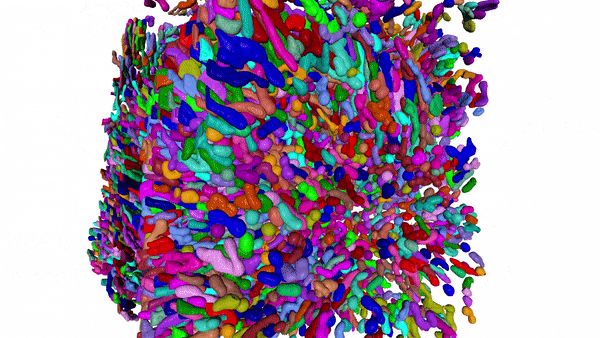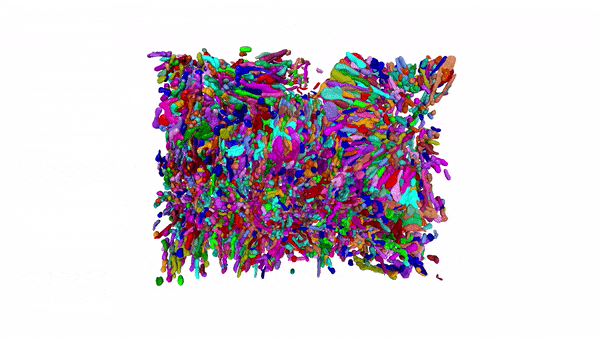
Our group has been developing and applying volume EM techniques to image cells and tissues; these include focused ion beam scanning electron microscopy (FIB-SEM) and more recently array tomography (AT), as well as other complementary methods. As the field of volume EM matures, the laboratory maintains an active technology development portfolio to interrogate an ever-widening variety of cell biological samples.
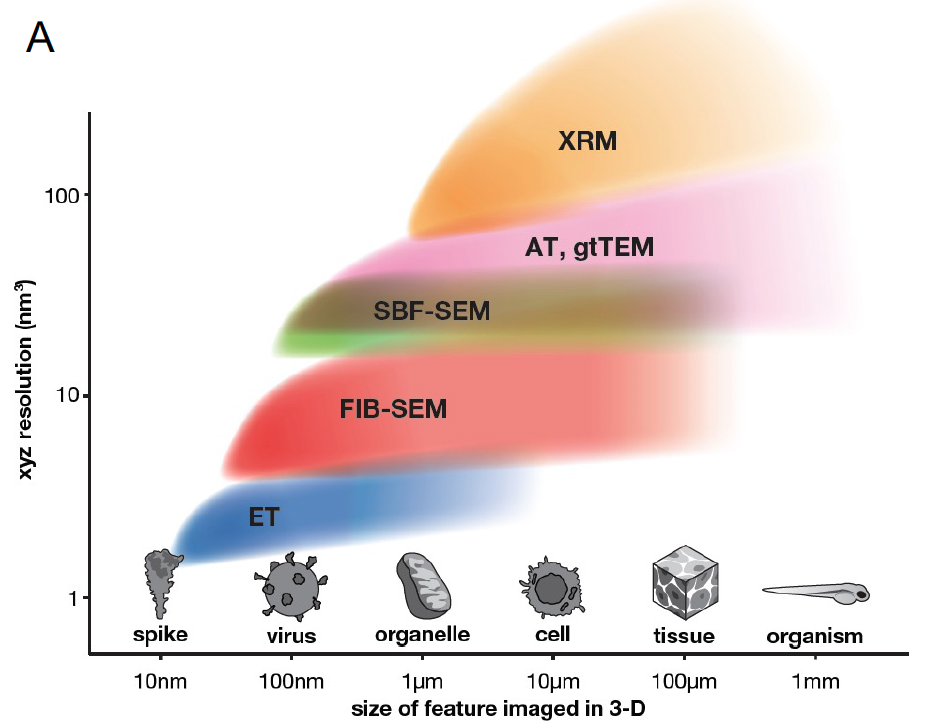
What is volume EM?
Several recently developed tools, including a variety of super-resolution light microscopy techniques and 3-D electron microscopy (EM) techniques, are capable of providing nanoscale information about cells and tissues. Traditionally, serial sectioning of resin-embedded cells and tissue samples followed by transmission EM (TEM) has been the approach of choice. More recently, the scanning electron microscope (SEM) is being paired with various sectioning approaches to generate near TEM-quality ultrastructural data. New hardware and software advances allow SEM-based approaches to imaging significantly larger volumes of the sample at high speed and with significant automation. Together, these imaging approaches that allow the interrogation of cells and tissue at µm to mm volume scales at nm level resolutions, we collectively refer to as volume EM techniques. As the term “3DEM” is increasingly applied broadly to high-resolution imaging-based structural biological studies, we use the term volume EM to refer to cell biological imaging studies that tackle significantly larger volumes.
At the CvEM, our primary techniques include focused ion beam scanning electron microscopy (FIB-SEM), often in conjunction with light or fluorescence microscopy, to image large samples in 3-D and at nanoscale resolutions. We have now employed FIB-SEM and CLEM/FIB-SEM in a variety of systems and continue to improve and expand this technology. In parallel, we are developing Array Tomography for samples that are significantly larger and that may not need the z-resolution that FIB-SEM affords.
For a primer on volume EM, see Baena V et al 2021 and Peddie CJ et al 2022
For a review on the use of focused ion beams in biology, see Narayan K and Subramaniam S 2015.
Visualizing cells in 3-D and at the nanoscale:
an introduction to focused ion beam scanning electron microscopy (FIB-SEM)
